Narrative review of radiomics for classifying pulmonary nodules and potential impact on lung cancer screening
Introduction
The advent of lung cancer screening has shown to reduce lung cancer mortality (1,2). Currently the process of lung cancer screening requires the identification of nodules on a chest computed tomography (CT) and subsequent risk stratification of those nodules. In the United States risk stratification is done primarily using LUNG-RADS and places nodules into categories ranging from 1–4 with subcategorization of category 4 into A, B, and X with each category showing increased risk for lung cancer (3). LUNG-RADS is based on well documented clinical observations of pulmonary nodules that show that size and density characteristics (solid, part-solid, and ground glass) are the most accurate imaging features that separate benign or malignant nodules. LUNG-RADS also includes sections that recommend tailoring recommendations based on features typical of intrapulmonary lymph nodes including subpleural location and triangular shape. While LUNG-RADS has proven successful, there is increasing interest in using both clinical and additional radiologic features to further improve risk stratification. Many of the radiologic features that have been proposed to achieve better categorization use computational methods to create the features and are often referred to a radiomic features. To better understand radiomic features and their potential impact on lung cancer screening this paper addresses the following questions: What are the traditional ways pulmonary nodules are currently classified? What are the most common radiomic features used for pulmonary nodule classification? How does the process of deriving radiomic features influence their utility? What is the performance of various radiomic features in their ability to better classify pulmonary nodules in the context of benign vs. malignant nodules, invasive vs. noninvasive adenocarcinomas, and recurrence risk following surgical resection. The author presents the following article in accordance with the Narrative Review reporting checklist (available at https://ccts.amegroups.com/article/view/10.21037/ccts-20-168/rc).
Methods
The primary literature search for this review was done on PubMed using the key terms of radiomics and lung cancer screening on September 1, 2020. In all, there were approximately 482 potential articles retrieved from the database. These articles were then reviewed and categorized into several different categories based on the clinical question they were trying to answer. Review articles were excluded from further consideration. In all 20 articles were found for evaluating factors that affect radiomic feature derivation and use, 28 articles were found evaluating radiomics in the context of classifying benign and malignant pulmonary nodules, 14 articles were found that evaluated radiomics to classify invasive and noninvasive adenocarcinomas, and 5 articles were found that used radiomics to try to predict recurrence risk following surgical resection.
Articles relating to traditional morphologic nodule features and clinical data for classifying pulmonary nodules are numerous and a selected group of key articles were selected to cover the breadth of proposed models for general overview purposes.
Articles related to specific radiomic features were found from reading the articles from the primary literature search.
Discussion
Traditional nodule features
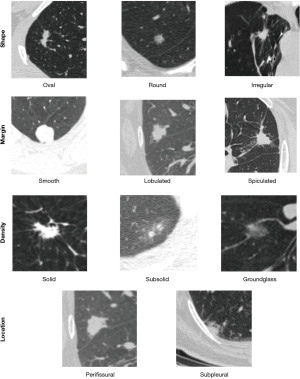
To understand radiomic features, it is important to review the traditional way radiologists have categorized pulmonary nodules and the limitations of this categorization. Until recently most research into pulmonary nodule classification required a radiologist to view a nodule and visually try to categorize it into common patterns. The easiest feature to include was the size of the nodule. It has been shown that size is the most reliable feature that differentiates benign from malignant nodules (4). Less robust features that also have shown differences between benign and malignant nodules include nodule density, border, shape, and location (5) with some attempt to formalize these features (6). A summary of these features is shown in Figure 1.
There are many observational studies that have explored these features as well as clinical features to try and improve the categorization of benign and malignant nodules (7-13), and concentrating on negative predictive value may reduce the overall number of repeat CT scans (14). The major limitation of these methods is that many of the features are subjective and left to the individual radiologist to correctly categorize each feature. The more features being considered the greater the chance for interobserver variability. It is primarily these limitations that have led to LUNG-RADS as being the preferred method. While LUNG-RADS does not include all possible features that may be important, it does create a framework for optimal reproducibility with relatively high sensitivity at the cost of lower specificity.
Radiomic nodule features
Radiomic nodule features are derived quantitative values from the nodule pixel data. These values can correlate to things like nodule location, nodule size, nodule radiodensity, nodule shape, nodule margin, nodule border, and nodule texture. Transform based features and features derived from unsupervised machine learning algorithms have also been considered when building models for classification. We will briefly describe some of the most common radiomic nodule features and how they are derived with a summary in Table 1.
Table 1
| Radiomic feature type | Description |
|---|---|
| Radiodensity | |
| Skewness | Measure of asymmetry |
| Kurtosis | Likelihood of an extreme value within the distribution |
| Uniformity | Similarity of the pixel values |
| Entropy | Randomness within the pixel values |
| Shape | |
| Sphericity | Quantifies how close to a sphere a given volume is |
| Compactness | Measures how well something fits into a defined shape |
| Margin | |
| Sharpness | Quantifies how different the pixels are at the nodule margin |
| Box counting fractal dimension | Quantifies complexity of the boarder |
| Second order texture features (GLCM) | |
| Homogeneity | How close the distribution of the values in the matrix are to the diagonal |
| Entropy | Measures texture irregularity |
| Uniformity | Measures how uniform the pixels are and equals 1 when all pixels are the same value |
| Contrast | Measures local intensity variation and is higher when there are more values away from the diagonal |
| Higher order texture features (GLSZM) | |
| Short zone emphasis | Measures pixel distribution of small zones and is higher for fine textures |
| Intensity non-uniformity | Measures similarity of pixel intensity throughout the image and is small if they are similar throughout the image |
| Zone percentage | Measures the homogeneity of zones in an image in a specific direction and is largest when the size of a zone is 1 for all intensities |
| Intensity variability | Measures the similarity of intensities throughout the image and is small if values are similar |
| Size zone variability | Measures similarity of the size of zones of various intensities throughout the image and is small when the sizes are similar |
| Higher order texture features (GLRLM) | |
| Short run emphasis | Measures distribution of short runs and is larger for fine textures |
| Grey-level non-uniformity | Measures the similarity of intensity values throughout the image and is small if intensity values are alike throughout the image |
| Run percentage | Measures the homogeneity and distribution of runs in a specific direction and is largest when the length of runs is 1 for all intensity values in the given direction |
| Intensity variability | Measures the similarity of intensity values throughout the image and is small if intensity values are alike throughout the image |
| Run length variability | Measures the similarity of the size of zones throughout the image and is small if the run lengths are alike throughout the image |
| Higher order texture features (NGTDM) | |
| Coarseness | Measures granularity within an image |
| Complexity | Measures the different intensity values throughout the image and is higher if there is a larger variety of values |
| Contrast | Measures intensity differences between neighbouring regions and is higher the more they differ |
| Busyness | Measures how much of a change in pixel intensity values there is from one pixel to the next and it is higher when changes are greater |
GLCM, grey level co-occurrence matrix; GLSZM, grey level size-zone matrix; GLRLM, grey level run length matrix; NGTDM, neighbourhood grey tone difference matrix.
Location features
Location features try to quantify the nodules location in relationship to a set point of the anatomy. For lung nodules, the minimum distance from the carina or pleural surface are most often starting points. In addition to the derived minimum distance, the distance can be decomposed into x, y, and z distances quantifying things such as whether or not the nodule is above or below the carina.
Size features
Nodule size features include volume, surface area, maximum diameter, minimum diameter, and average diameter. To derive these values requires accurate segmentation of the nodule.
Radiodensity features
Features related to nodule radiodensity encompass those features that describe the overall distribution of the pixel data within the nodule. These features are also referred to as first-order statistics in texture analysis as they are only considering each pixel values in isolation and give a global picture of the pixel data (15). They most often include the mean pixel value, median pixel value, maximum pixel value, minimum pixel value, skewness, kurtosis, entropy, and uniformity. While the mean, median, maximum, and minimum pixel value are self-explanatory, we will briefly describe the other features.
Skewness is the measure of asymmetry and deviation from the normal distribution. A skewness of zero implies a normal distribution. A skewness greater than 0 implies the distribution is skewed to the right (more values that have higher intensity) while a skewness less than 0 implies the distribution is skewed to the left (more values with lower intensity values).
Kurtosis describes the peakedness of the distribution. If the kurtosis is three then the distribution is a normal distribution. If the kurtosis is greater than three, it implies that it is a sharper distribution with values concentrated around the mean but with thicker tails meaning there is a higher probability of an extreme value. If kurtosis is less than three the distribution is flatter with a wider peak meaning there are less extreme values but the range is spread out more broadly around the mean.
Entropy is a measure of texture randomness or irregularity. The greater the entropy the more heterogeneous and varied the pixel data. Conversely uniformity is the sum of the squared elements in the region of interest (ROI) and takes on higher values when the pixels have a similar value.
Shape features
There are many methods that can be used to describe a nodules shape. The most common features used are sphericity (16) and compactness (17). More details about additional shape features are discussed in a review article by Yang et al. (18).
Margin features
Margin features attempt to quantify the subjective radiologist classification of a nodule as being either smooth, irregular, or spiculated. Box counting fractal analysis is a common method for quantifying irregularity of a shapes margin using the fractal dimension (19). The most commonly used fractal method uses a box counting method with higher values correlating to more complicated margins. Methods for looking at margin sharpness have also been implemented looking at the distribution of pixel values along the margins and deriving first-order texture features on the pixel values.
Second and higher order texture features
Second order and higher order texture features are features that take into account more than one pixel. Second-order statistics take into account one additional pixel a defined distance and angle from each pixel in the ROI. The algorithm then counts co-occurrences of pixels with similar values for each angle and create what are called a grey-length co-occurrence matrices (GLCM) for each angle and uses it to calculate features related to heterogeneity including homogeneity, uniformity, contrast, and entropy (20). Homogeneity will be large if the co-occurrences cluster along the diagonal of the matrix while contrast will be large if the co-occurrences cluster away from the diagonal. Images with large values for contrast in a given direction implies that there are increased linear structures when going that direction. Uniformity measures the sum of squared elements in the matrix. It takes a value between 0 and 1 and is 1 when all pixels have similar values and co-occur together. Entropy is a measure of randomness. Inhomogeneous textures have low entropy while homogenous textures will have high entropy. Changes in defined distance, angles being considered, and how you define when pixels co-occur all contribute to the features being produced, and as such many more second order features are usually considered compared to their first-order counterparts.
Higher order texture features consider three or more pixels in relation to another pixel. The three most common algorithms used create a grey level size-zone matrix (GLSZM), a grey level run length matrix (GLRLM), or a neighborhood grey tone difference matrix (NGTDM) (21-23). GLSZM considers regional variations and produces values such as short zone emphasis, intensity non-uniformity, zone percentage, intensity variability, and size zone variability. GLRLM also considers regional variations and produces features such as short run emphasis, grey-level non-uniformity, run percentage, intensity variability, and run length variability. NGTDM considers more local variations and produces features such as coarseness, complexity, contrast, and busyness. Because of their complexity higher order texture features tend to have more variables that can be set and can produce a larger number of potential features to consider.
Transformed-based features
Transformed-based features most often use a wavelet transform to capture both frequency and location information in order to extract certain levels of noise to enhance potential textures within an image that may be relevant. First, second, and higher order textural analysis can then be applied to the transformed image. Given the additional filtering, these methods can produce an even greater number of features for a given ROI.
Features derived from unsupervised machine learning
Unsupervised machine learning algorithms can be used to find common patterns among nodules. These methods are often referred to as clustering algorithms and are designed to group data having similar patterns. When applying these algorithms to nodules, nodule ROI pixel data can be divided into smaller blocks. Blocks can then be compared using these methods and grouped depending on similarity into groups called clusters. Presence of a block belonging to a particular cluster can then be used as a feature for classification. An example of such a method has been used to differentiate between EGFR subtypes (24).
Perinodular features
Perinodular features use the pixel data surrounding the nodule ROI to calculate texture features, clustering features, and transformed-based features. These features can then be used with the nodule features for potentially more robust classification of nodules.
Dynamic features
Dynamic radiomic features are features derived from changes in radiomic feature over time. This may be applicable for multiphase contrast studies or follow up imaging of a nodule.
Variation in radiomic nodule features
To derive radiomic nodule features there are many steps that must take place (Figure 2). This section will briefly describe each of these steps and the parameters that can lead to variation in the different nodule features.
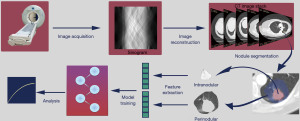
Acquisition of CT
When a CT is acquired there are many variable parameters that must be set that determine the time the scan takes and the amount of noise that will be produced. The primary parameters that affect these outcomes include mA, KvP, and gantry. Changing these parameters can create differences in background noise and may artifactually increase or decrease values of derived texture parameters. Fixed parameters can also cause variations in the background noise of the scan and thus radiomic feature variation. These parameters include differences in scanner models, vendor of the CT scanner, size of the CT scanner, and the size of the patient. It has been shown that derived texture features can vary across scanners often grouping together based on the CT scanner rather than the actual texture (25). In addition, breathing phase and mA have also shown significant variation among radiomic features (26). Methods have been proposed to reduce this variation and find more robust features (27). It has also been shown that scanning during certain parts of the breathing cycle may reduce variability (28).
Reconstruction and postprocessing of CT images
When a CT is performed it generates what is called a sinogram. This is the raw data produced by the CT scanner. In order for the CT scanner to produce images it must decompose the sinogram. Depending on the algorithm used this can result it images of various slice thicknesses and noise. The algorithm used to produce an image is usually referred to as a kernel and differences in the kernel can change the values of texture features. How much noise an image has will play a role in derived texture features. Using different kernels and filter sizes have shown to cause variation in derived texture features (29). It has also been shown radiomic features can be affected by slice thickness and reconstruction algorithms (30,31). Some work has been done to adjust for this noise when comparing different kernels (32).
Nodule segmentation
Once images are obtained the nodules must be identified and parsed from the image. For most studies this is a manual process performed by a radiologist who is sometimes assisted by a semi-automated segmentation algorithm. This can lead to variability predominantly in radiomic features relating to shape and border (33). Semiautomatic volumetric segmentation has been shown to have better correlation among extracted radiomic features than manual segmentation (34) and differences in segmentation algorithms have been shown to effect consistency of extracted textual features (35). Recent advances in deep neural networks have the potential to fully automate segmentation in the near future and hopefully reducing this type of variation (36,37).
Radiomic feature derivation
Depending on the radiomic feature there may be parameters that are set by the user that can have an effect on the derived value. This is particularly true for second and higher order texture features, transform based features, and features derived from unsupervised machine learning algorithms. How intensity values of CT and positron emission tomography (PET) data is grouped together (aka binning) has been shown to have significant effects in the variability of texture features with optimal number of intensity values of 128 showing the least variability (38,39). Radiomic texture features derived from GLCM may be more robust compared to features derived from other matrices (40). Shape features can be affected by a nodule size (41). Welch et al. also showed tumor size can create variation in derived radiomic features. To limit such sources of variation they propose to use open-source software to increase accountability and reproducibility, compare features to existing morphologic features such as tumor volume, compensate for overlapping features, evaluate features for dependencies using statistical analysis, and pre-process data to ensure good image signal quality (42). There has also been work showing dynamic radiomic texture features may be more robust than texture features derived from a single time point (43).
Statistical analysis
The analysis done to evaluate the performance of radiomic features can have a significant impact on the perceived clinical usefulness. For example, Kaplan-Meier survival curves and associated P values can vary widely depending on how cutoff values are set for derived radiomic features (44).
Radiologist compliance
Using texture features in clinical decision making will require the radiologist to be fully involved. Miles et al. found that when implementing texture features in routine clinical practice that radiologists only showed 59% compliance decreasing with time. User friendly software, ease of incorporation into reports, and seamless integration into PACS all played a role in compliance (45).
Performance of radiomic nodule features
There have been many proposed applications for texture features as it relates to evaluating pulmonary nodules. The primary application for lung cancer screening is differentiating benign and malignant nodules. In addition, differentiating between invasive and noninvasive adenocarcinomas and predicting surgical recurrence risk in patients with confirmed cancer are also active areas of research that could impact lung cancer screening workflows. In this section we will summarize the current research relating to these potential applications.
Predicting benign vs. malignant nodules
Research related to distinguishing benign and malignant pulmonary nodules using radiomic features has exploded over the past ten years. The majority of studies are retrospective studies without independent data sets for validation. This makes comparison between methods difficult as there is no standard data set for comparison. Most studies also do not compare their accuracy with existing methods that do not use radiomic features, also complicating assessment of their true value.
Initial work exploring texture features for distinguishing benign and malignant nodules were limited to small data sets and lack of validation. These studies explored primarily first-order texture features, second-order texture features, and fractal analysis. Analysis was primarily focused on whether there was a significant difference between the benign and malignant group for a given feature and did not formally evaluate classification performance. Entropy and correlation were initial features found to show a difference between the two groups (46,47). An early study on fractal analysis suggested values derived from 2D and 3D box counting methods of the binary and grey-scale pixel data showed significant differences between benign and malignant nodules as well as between adenocarcinoma and squamous cell carcinoma (48).
Following these initial studies, a variety of radiomic features were explored to create prediction models. Studies using only first-order texture features using small data sets appeared to show conflicting features that were important with mean attenuation and entropy being features that showed differences over multiple studies (49-51). There were no validation sets and area under the curve (AUC) were high, ranging between 0.864–0.93. A more recent study looking at only kurtosis and skewness showed differences between benign and malignant nodules; however, AUC was only approximately 0.7 for both features and no combined model was explored (52).
Some early studies also looked at derived features from unsupervised methods. For example, features derived from principle component analysis on a large number of manually segmented benign and malignant nodules were filtered and used to train a support vector machine classifier showing AUC of 0.908 on the training set (53).
Studies using additional radiomic features outside of texture are now more common. In addition, the number of texture features being considered are also increasing due to wavelet transforms (54). While many of these use their own unique data sets, several use a standard database of nodules from the LIDC dataset. Models using this standard data set had AUCs ranging from 0.927–0.97 with the highest using shape and texture features in conjunction with random forests (55-58). The model having the highest AUC combined shape features, texture features, and features derived from a deep convolution neural network (58). A simple model using only margin and texture features was able to achieve an AUC of 0.856 using a random forest tree method. They also showed that features derived from statistical models were better than features derived from simple correlation (59).
There has been some work in incorporating perinodular texture features into prediction models for both benign and malignant nodules. One of these models showed good performance on both training and validation training sets after clustering features to create common centroid features that could then be used to train a neural network. They were able to get an AUC of 0.965 on an independent data set as compared to 0.594 using diameter alone (60). An additional model used to distinguish between malignant nodules and granulomas used intra and perinodular texture features to train a support vector machine. The model had an AUC of 0.8 on an independent dataset and improved classification performance compared to a model only using intramodular texture features and a deep neural network trained on the whole nodule volume (61).
While most studies did not compare the performance of their methods to traditional methods such as LUNG-RADS there has been work that compared the two. Using a variety of radiomic features Mao et al. created a statistical model that had a sensitivity of 0.8 and specificity of 0.9 compared to a sensitivity of 0.9 and specificity of 0.77 for LUNG-RADS (62). This study raises the possibility that these models may add specificity to increase performance at the cost of lost sensitivity.
There have also been recent studies that have broken down what features are most reproducible and useful for classification. One suggests that first order statistics and NGLDM do not produce reproducible features that can distinguish between benign and malignant nodules. Its methods favored texture features derived from GLCM and GLRLM (63). It is difficult to know how accurate this finding is given there was no description of the three data sets they used for extracting features, training their model, and validating their model. An additional study broke down features found to be reproducible and able to distinguish between benign and malignant nodules. They found 14 of 330 GLCM, 1 of 49 first-order features, and 5 or 18 shape features to fit their criteria. They found that a model using shape features performed the best (64). While the studies are limited, they seem to suggest first order statistics may not have a significant role to play in differentiating benign and malignant nodules compared to features derived from shape and texture features derive from second- and higher-order statistics.
Predicting noninvasive and invasive adenocarcinomas
There have been several studies exploring the use of radiomic features to distinguish invasive and noninvasive adenocarcinomas in ground glass and part-solid nodules. Much of this work primarily focused on morphologic features and first- and second-order texture features (65-72). AUCs of these studies ranged from 0.78–0.98. The features found to be significantly different between the two groups varied greatly with size features such as volume and texture features such as entropy being the most common showing differences between the two groups. Generally, it was found that nodules with higher entropy and lower homogeneity appeared to be more associated with invasive adenocarcinomas. One of the studies found that adding simple texture features did not improve performance compared to simply using the percent solid component and size which raises the possibility there may not be much added value to using these low level texture features compared to current practice guidelines that suggest following changes in the solid component of the nodule (71).
A more recent study looking at texture, morphologic, and clinical data for both 2D ROIs and 3D VOIs found that maximum diameter and spiculation were the best morphologic features and that only 11 of 1,125 texture features were found to be significant. There best performing model used the 2D features only with AUC of 0.938 compared to 0.753 for a 3D based feature model. Clinical data did not appear to add any value to the model (73).
There has also been work using machine learning derived features for classification of invasive and noninvasive adenocarcinomas for both nonenhanced and enhanced chest CTs that showed a maximum AUC of 0.89. There was not a significant difference between models using features derived from the unenhanced and enhanced CTs (74).
Combining radiomic features and deep learning machine learning has also been studied showing an AUC of 0.966 when combining methods while demonstrating an AUC of 0.744 and 0.847 using deep learning and radiomic features respectively (75).
Predicting surgical recurrence risk
There are a small number of studies looking at using radiomic features to predict recurrence risk of surgically resected tumors. Akinci et al. extracted morphological, first-order statistical, higher-level textural features, and fractal-based features from the gross tumor volume and peritumoral volume. They used these features to predict tumor recurrence in stage IA and IIB tumors that were resected. They then compared the predictive value of their model to the typical TNM staging and found it improved performance from an AUC of 0.68 to and AUC of 0.76 (76).
Depeursinge et al. looked at a group of 101 patients with surgically resected stage I adenocarcinomas and created a model using wavelet-based features from the entire nodule or only the portion containing a solid component. They found that using the solid component only improved the overall performance of the model with the best model achieving an AUC of 0.81 (77).
There are several studies looking at primarily texture features to try and predict tumor recurrence risk following surgery for early stage NLCLC from CT and PET pixel intensities. One study with a small sample size found gray-level correlation and sum variance tended to be higher in the local recurrence group (78). A larger study of 194 patients created a logistic regression model using extracted texture features at both 1 and 5 mm CT slice thickness and found that skewness and kurtosis were poorly correlated between the different slice thicknesses while entropy and mean attenuation were highly correlated. Overall their model was able to achieve and AUC of 0.81 (79). A small study of 55 patients looking at FDG-PET data found that tumor stage and intensity variability were the only independent predictors of progression free survival after multivariate analysis. They did not attempt to build a predictive model (80).
Summary
The use of computational nodule features has the potential to improve accuracy of lung cancer screening and subsequent decision making of lung cancer management however there are still many hurdles to overcome due to variabilities in derived radiomic features secondary to differences in imaging acquisition, image reconstruction, nodule segmentation, and algorithm parameters used for computing various radiomic nodule features. While efforts are being made to standardize workflows and methods to extract radiomic features, there has been no widespread adoption of any particular feature. Institutions interested in using these features currently would need to standardize their own workflows and establish features that they feel are reliable in the setting of their institution.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Sandra Starnes) for the series “Lung Cancer Screening” published in Current Challenges in Thoracic Surgery. The article has undergone peer review.
Reporting Checklist: The author has completed the Narrative Review reporting checklist. Available at https://ccts.amegroups.com/article/view/10.21037/ccts-20-168/rc
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://ccts.amegroups.com/article/view/10.21037/ccts-20-168/coif). The series “Lung Cancer Screening” was commissioned by the editorial office without any funding or sponsorship. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Patz EF, Caporaso NE, Dubinett SM, et al. National Lung Cancer Screening Trial American College of Radiology Imaging Network Specimen Biorepository originating from the Contemporary Screening for the Detection of Lung Cancer Trial (NLST, ACRIN 6654): design, intent, and availability of specimens for validation of lung cancer biomarkers. J Thorac Oncol 2010;5:1502-6. [Crossref] [PubMed]
- de Koning HJ, van der Aalst CM, de Jong PA, et al. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N Engl J Med 2020;382:503-13. [Crossref] [PubMed]
- Lung Rads [Internet]. [cited 2020 Sep 17]. Available online: https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/Lung-Rads
- Henschke CI, Yankelevitz DF, Naidich DP, et al. CT screening for lung cancer: suspiciousness of nodules according to size on baseline scans. Radiology 2004;231:164-8. [Crossref] [PubMed]
- Ko JP. Lung nodule detection and characterization with multi-slice CT. J Thorac Imaging 2005;20:196-209. [Crossref] [PubMed]
- Farag A, Elhabian S, Graham J, et al. Toward precise pulmonary nodule descriptors for nodule type classification. Med Image Comput Comput Assist Interv 2010;13:626-33.
- Gurney JW. Determining the likelihood of malignancy in solitary pulmonary nodules with Bayesian analysis. Part I. Theory. Radiology 1993;186:405-13. [Crossref] [PubMed]
- Swensen SJ, Silverstein MD, Ilstrup DM, et al. The probability of malignancy in solitary pulmonary nodules. Application to small radiologically indeterminate nodules. Arch Intern Med 1997;157:849-55.
- Herder GJ, van Tinteren H, Golding RP, et al. Clinical prediction model to characterize pulmonary nodules: validation and added value of 18F-fluorodeoxyglucose positron emission tomography. Chest 2005;128:2490-6. [Crossref] [PubMed]
- Gould MK, Ananth L, Barnett PG, et al. A clinical model to estimate the pretest probability of lung cancer in patients with solitary pulmonary nodules. Chest 2007;131:383-8. [Crossref] [PubMed]
- Li Y, Wang J. A mathematical model for predicting malignancy of solitary pulmonary nodules. World J Surg 2012;36:830-5. [Crossref] [PubMed]
- McWilliams A, Tammemagi MC, Mayo JR, et al. Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med 2013;369:910-9. [Crossref] [PubMed]
- Soardi GA, Perandini S, Motton M, et al. Assessing probability of malignancy in solid solitary pulmonary nodules with a new Bayesian calculator: improving diagnostic accuracy by means of expanded and updated features. Eur Radiol 2015;25:155-62. [Crossref] [PubMed]
- Stephens MJ, Rho BH, Curran-Everett D, et al. Identification of Nonaggressive Pulmonary Nodules Using an Optimized Scoring System. J Thorac Imaging 2019;34:170-8. [Crossref] [PubMed]
- Tomita T, Tsuji S. Computer Analysis of Visual Textures. Springer Science, 1990.
- Wadell H. Volume, Shape, and Roundness of Quartz Particles. J Geol 1935;43:250-80.
- Montero R, Bribiesca E. State of the Art of Compactness and Circularity Measures 1 [cited 2020 Sep 26]. Available online: /paper/State-of-the-Art-of-Compactness-and-Circularity-1-Montero-Bribiesca/8364d027fcaea36786472f1a71e9e0a9ea2b5298
- Yang M, Kpalma K, Ronsin J. A Survey of Shape Feature Extraction Techniques. Pattern Recognit 2008;43-90.
-
Fractal Geometry Kenneth F. 2nd - Haralick RM, Shanmugam K, Dinstein I. Textural Features for Image Classification. IEEE Trans Syst Man Cybern 1973;SMC-3:610-21.
- Thibault G, Fertil B, Navarro C, et al. Texture indexes and gray level size zone matrix. Application to cell nuclei classification. Pattern Recognit Inf Process 2009;140-5.
- Galloway MM. Texture analysis using gray level run lengths. Comput Graph Image Process 1975;4:172-9.
- Amadasun M, King R. Textural features corresponding to textural properties. IEEE Trans Syst Man Cybern 1989;19:1264-74.
- Clay R, Kipp BR, Jenkins S, et al. Computer-Aided Nodule Assessment and Risk Yield (CANARY) may facilitate non-invasive prediction of EGFR mutation status in lung adenocarcinomas. Sci Rep 2017;7:17620. [Crossref] [PubMed]
- Mackin D, Fave X, Zhang L, et al. Measuring Computed Tomography Scanner Variability of Radiomics Features. Invest Radiol 2015;50:757-65. [Crossref] [PubMed]
- Fave X, Cook M, Frederick A, et al. Preliminary investigation into sources of uncertainty in quantitative imaging features. Comput Med Imaging Graph 2015;44:54-61. [Crossref] [PubMed]
- Orlhac F, Frouin F, Nioche C, et al. Validation of A Method to Compensate Multicenter Effects Affecting CT Radiomics. Radiology 2019;291:53-9. [Crossref] [PubMed]
- Tanaka S, Kadoya N, Kajikawa T, et al. Investigation of thoracic four-dimensional CT-based dimension reduction technique for extracting the robust radiomic features. Phys Med 2019;58:141-8. [Crossref] [PubMed]
- Ketabi A, Ghafarian P, Mosleh-Shirazi MA, et al. Impact of image reconstruction methods on quantitative accuracy and variability of FDG-PET volumetric and textural measures in solid tumors. Eur Radiol 2019;29:2146-56. [Crossref] [PubMed]
- Kim YJ, Lee HJ, Kim KG, et al. The Effect of CT Scan Parameters on the Measurement of CT Radiomic Features: A Lung Nodule Phantom Study. Comput Math Methods Med 2019;2019:8790694. [Crossref] [PubMed]
- Lu L, Ehmke RC, Schwartz LH, et al. Assessing Agreement between Radiomic Features Computed for Multiple CT Imaging Settings. PLoS One 2016;11:e0166550. [Crossref] [PubMed]
- Choe J, Lee SM, Do KH, et al. Deep Learning-based Image Conversion of CT Reconstruction Kernels Improves Radiomics Reproducibility for Pulmonary Nodules or Masses. Radiology 2019;292:365-73. [Crossref] [PubMed]
- Pavic M, Bogowicz M, Würms X, et al. Influence of inter-observer delineation variability on radiomics stability in different tumor sites. Acta Oncol 2018;57:1070-4. [Crossref] [PubMed]
- Parmar C, Rios Velazquez E, Leijenaar R, et al. Robust Radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One 2014;9:e102107. [Crossref] [PubMed]
- Bashir U, Azad G, Siddique MM, et al. The effects of segmentation algorithms on the measurement of 18F-FDG PET texture parameters in non-small cell lung cancer. EJNMMI Res 2017;7:60. [Crossref] [PubMed]
- Qin Y, Zheng H, Huang X, et al. Pulmonary nodule segmentation with CT sample synthesis using adversarial networks. Med Phys 2019;46:1218-29. [Crossref] [PubMed]
- Rocha J, Cunha A, Mendonça AM. Conventional Filtering Versus U-Net Based Models for Pulmonary Nodule Segmentation in CT Images. J Med Syst 2020;44:81. [Crossref] [PubMed]
- Wang HYC, Donovan EM, Nisbet A, et al. The stability of imaging biomarkers in radiomics: a framework for evaluation. Phys Med Biol 2019;64:165012. [Crossref] [PubMed]
- Leijenaar RTH, Nalbantov G, Carvalho S, et al. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep 2015;5:11075. [Crossref] [PubMed]
- Cook GJR, Siddique M, Taylor BP, et al. Radiomics in PET: principles and applications. Clin Transl Imaging 2014;2:269-76.
- Niehaus R, Raicu DS, Furst J, et al. Toward Understanding the Size Dependence of Shape Features for Predicting Spiculation in Lung Nodules for Computer-Aided Diagnosis. J Digit Imaging 2015;28:704-17. [Crossref] [PubMed]
- Welch ML, McIntosh C, Haibe-Kains B, et al. Vulnerabilities of radiomic signature development: The need for safeguards. Radiother Oncol 2019;130:2-9. [Crossref] [PubMed]
- Nardone V, Reginelli A, Guida C, et al. Delta-radiomics increases multicentre reproducibility: a phantom study. Med Oncol 2020;37:38. [Crossref] [PubMed]
- McQuaid S, Scuffham J, Alobaidli S, et al. Factors influencing the robustness of P-value measurements in CT texture prognosis studies. Phys Med Biol 2017;62:5403-16. [Crossref] [PubMed]
- Miles KA, Squires J, Murphy M. Radiologist Engagement as a Potential Barrier to the Clinical Translation of Quantitative Imaging for the Assessment of Tumor Heterogeneity. Acad Radiol 2018;25:935-42. [Crossref] [PubMed]
- McNitt-Gray MF, Hart EM, Wyckoff N, et al. A pattern classification approach to characterizing solitary pulmonary nodules imaged on high resolution CT: preliminary results. Med Phys 1999;26:880-8. [Crossref] [PubMed]
- Wang H, Guo XH, Jia ZW, et al. Multilevel binomial logistic prediction model for malignant pulmonary nodules based on texture features of CT image. Eur J Radiol 2010;74:124-9. [Crossref] [PubMed]
- Kido S, Kuriyama K, Higashiyama M, et al. Fractal analysis of small peripheral pulmonary nodules in thin-section CT: evaluation of the lung-nodule interfaces. J Comput Assist Tomogr 2002;26:573-8. [Crossref] [PubMed]
- Lee SH, Lee SM, Goo JM, et al. Usefulness of texture analysis in differentiating transient from persistent part-solid nodules (PSNs): a retrospective study. PLoS One 2014;9:e85167. [Crossref] [PubMed]
- Suo S, Cheng J, Cao M, et al. Assessment of Heterogeneity Difference Between Edge and Core by Using Texture Analysis: Differentiation of Malignant From Inflammatory Pulmonary Nodules and Masses. Acad Radiol 2016;23:1115-22. [Crossref] [PubMed]
- Dennie C, Thornhill R, Sethi-Virmani V, et al. Role of quantitative computed tomography texture analysis in the differentiation of primary lung cancer and granulomatous nodules. Quant Imaging Med Surg 2016;6:6-15. [Crossref] [PubMed]
- Borguezan BM, Lopes AJ, Saito EH, et al. Solid Indeterminate Nodules with a Radiological Stability Suggesting Benignity: A Texture Analysis of Computed Tomography Images Based on the Kurtosis and Skewness of the Nodule Volume Density Histogram. Pulm Med 2019;2019:4071762. [Crossref] [PubMed]
- Wang JJ, Wu HF, Sun T, et al. Prediction models for solitary pulmonary nodules based on curvelet textural features and clinical parameters. Asian Pac J Cancer Prev 2013;14:6019-23. [Crossref] [PubMed]
- Gao N, Tian S, Li X, et al. Three-Dimensional Texture Feature Analysis of Pulmonary Nodules in CT Images: Lung Cancer Predictive Models Based on Support Vector Machine Classifier. J Digit Imaging 2020;33:414-22. [Crossref] [PubMed]
- Han F, Wang H, Zhang G, et al. Texture feature analysis for computer-aided diagnosis on pulmonary nodules. J Digit Imaging 2015;28:99-115. [Crossref] [PubMed]
- Wu W, Hu H, Gong J, et al. Malignant-benign classification of pulmonary nodules based on random forest aided by clustering analysis. Phys Med Biol 2019;64:035017. [Crossref] [PubMed]
- Xie Y, Zhang J, Xia Y. Semi-supervised adversarial model for benign-malignant lung nodule classification on chest CT. Med Image Anal 2019;57:237-48. [Crossref] [PubMed]
- Zhang G, Yang Z, Gong L, et al. Classification of benign and malignant lung nodules from CT images based on hybrid features. Phys Med Biol 2019;64:125011. [Crossref] [PubMed]
- Ferreira JR, Oliveira MC, de Azevedo-Marques PM. Characterization of Pulmonary Nodules Based on Features of Margin Sharpness and Texture. J Digit Imaging 2018;31:451-63. [Crossref] [PubMed]
- Uthoff J, Stephens MJ, Newell JD, et al. Machine learning approach for distinguishing malignant and benign lung nodules utilizing standardized perinodular parenchymal features from CT. Med Phys 2019;46:3207-16. [Crossref] [PubMed]
- Beig N, Khorrami M, Alilou M, et al. Perinodular and Intranodular Radiomic Features on Lung CT Images Distinguish Adenocarcinomas from Granulomas. Radiology 2019;290:783-92. [Crossref] [PubMed]
- Mao L, Chen H, Liang M, et al. Quantitative radiomic model for predicting malignancy of small solid pulmonary nodules detected by low-dose CT screening. Quant Imaging Med Surg 2019;9:263-72. [Crossref] [PubMed]
- Yan M, Wang W. A Non-invasive Method to Diagnose Lung Adenocarcinoma. Front Oncol 2020;10:602. [Crossref] [PubMed]
- Liu C, Ma C, Duan J, et al. Using CT texture analysis to differentiate between peripheral lung cancer and pulmonary inflammatory pseudotumor. BMC Med Imaging 2020;20:75. [Crossref] [PubMed]
- Chae HD, Park CM, Park SJ, et al. Computerized texture analysis of persistent part-solid ground-glass nodules: differentiation of preinvasive lesions from invasive pulmonary adenocarcinomas. Radiology 2014;273:285-93. [Crossref] [PubMed]
- Son JY, Lee HY, Lee KS, et al. Quantitative CT analysis of pulmonary ground-glass opacity nodules for the distinction of invasive adenocarcinoma from pre-invasive or minimally invasive adenocarcinoma. PLoS One 2014;9:e104066. [Crossref] [PubMed]
- Hwang IP, Park CM, Park SJ, et al. Persistent Pure Ground-Glass Nodules Larger Than 5 mm: Differentiation of Invasive Pulmonary Adenocarcinomas From Preinvasive Lesions or Minimally Invasive Adenocarcinomas Using Texture Analysis. Invest Radiol 2015;50:798-804. [Crossref] [PubMed]
- Yagi T, Yamazaki M, Ohashi R, et al. HRCT texture analysis for pure or part-solid ground-glass nodules: distinguishability of adenocarcinoma in situ or minimally invasive adenocarcinoma from invasive adenocarcinoma. Jpn J Radiol 2018;36:113-21. [Crossref] [PubMed]
- Sun Q, Huang Y, Wang J, et al. Applying CT texture analysis to determine the prognostic value of subsolid nodules detected during low-dose CT screening. Clin Radiol 2019;74:59-66. [Crossref] [PubMed]
- Cohen JG, Reymond E, Medici M, et al. CT-texture analysis of subsolid nodules for differentiating invasive from in-situ and minimally invasive lung adenocarcinoma subtypes. Diagn Interv Imaging 2018;99:291-9. [Crossref] [PubMed]
- Alpert JB, Rusinek H, Ko JP, et al. Lepidic Predominant Pulmonary Lesions (LPL): CT-based Distinction From More Invasive Adenocarcinomas Using 3D Volumetric Density and First-order CT Texture Analysis. Acad Radiol 2017;24:1604-11. [Crossref] [PubMed]
- She Y, Zhang L, Zhu H, et al. The predictive value of CT-based radiomics in differentiating indolent from invasive lung adenocarcinoma in patients with pulmonary nodules. Eur Radiol 2018;28:5121-8. [Crossref] [PubMed]
- Yang G, Nie P, Zhao L, et al. 2D and 3D texture analysis to predict lymphovascular invasion in lung adenocarcinoma. Eur J Radiol 2020;129:109111. [Crossref] [PubMed]
- Gao C, Xiang P, Ye J, et al. Can texture features improve the differentiation of infiltrative lung adenocarcinoma appearing as ground glass nodules in contrast-enhanced CT? Eur J Radiol 2019;117:126-31. [Crossref] [PubMed]
- Wang X, Zhang L, Yang X, et al. Deep learning combined with radiomics may optimize the prediction in differentiating high-grade lung adenocarcinomas in ground glass opacity lesions on CT scans. Eur J Radiol 2020;129:109150. [Crossref] [PubMed]
- Akinci D’Antonoli T, Farchione A, Lenkowicz J, et al. CT Radiomics Signature of Tumor and Peritumoral Lung Parenchyma to Predict Non-small Cell Lung Cancer Postsurgical Recurrence Risk. Acad Radiol 2020;27:497-507. [Crossref] [PubMed]
- Depeursinge A, Yanagawa M, Leung AN, et al. Predicting adenocarcinoma recurrence using computational texture models of nodule components in lung CT. Med Phys 2015;42:2054-63. [Crossref] [PubMed]
- Dennie C, Thornhill R, Souza CA, et al. Quantitative texture analysis on pre-treatment computed tomography predicts local recurrence in stage I non-small cell lung cancer following stereotactic radiation therapy. Quant Imaging Med Surg 2017;7:614-22. [Crossref] [PubMed]
- Koo HJ, Sung YS, Shim WH, et al. Quantitative Computed Tomography Features for Predicting Tumor Recurrence in Patients with Surgically Resected Adenocarcinoma of the Lung. PLoS One 2017;12:e0167955. [Crossref] [PubMed]
- Nakajo M, Jinguji M, Shinaji T, et al. A Pilot Study of Texture Analysis of Primary Tumor [18F]FDG Uptake to Predict Recurrence in Surgically Treated Patients with Non-small Cell Lung Cancer. Mol Imaging Biol 2019;21:771-80.
Cite this article as: Stephens MJ. Narrative review of radiomics for classifying pulmonary nodules and potential impact on lung cancer screening. Curr Chall Thorac Surg 2023;5:4.